
Kristen Kroll, PhD
Professor, WashU Developmental Biology
- Phone: 314-747-5519
- Email: kkroll@nospam.wustl.edu
Transcriptional and epigenetic regulation of neural development
Our research focuses on defining regulatory networks controlling early neuroectodermal specification, later neurogenesis, and the generation of particular neuronal cell types. We are particularly interested in understanding how epigenetic regulation modulates these networks and how epigenome dysregulation contributes to neurodevelopmental disorders, including epileptic encephalopathies, neural tube defects, and pediatric and adult glioblastoma. This work uses directed differentiation of human and mouse embryonic stem cells (ESCs), human primary tumor cell lines, and mouse models, in combination with genome-wide approaches, to assess how transcriptome and chromatin state changes are regulated during developmental transitions.

Gene regulatory networks in embryonic neural progenitor cells. (Figure A-D) In prior work, we defined transcriptional programs and enhancers controlled by two neural bHLH transcription factors during neurogenesis and roles for epigenetic regulatory activities, including the Polycomb complex and the nucleoprotein Geminin, in epigenetic control of several aspects of embryonic cell specification. More recently, we defined direct targets of Geminin and another neural regulatory transcription factor by ChIP-seq analysis in embryonic neural progenitor cells. By combining these and additional data for multiple neural regulatory transcription factors, we are constructing a gene regulatory network (GRN) of transcriptional and epigenetic regulatory activities that are direct targets of neural-regulatory transcription factors in telencephalic neural progenitor cells (Figure A-D). We are characterizing how disruption of this network contributes to the etiology of neural tube defects (NTDs). Although these are the second most common birth defect (~1:1000 fetuses and newborns) their causation is poorly understood. This is in part because NTD occurrence usually has a multi-factorial etiology, with both genetic and environmental susceptibility factors contributing to phenotypic manifestation and severity. Defining how this network is altered in different NTD models may indicate core processes that, when disrupted, increase NTD susceptibility or serve as early biomarkers. Through this work we are also defining new epigenetic regulators with likely roles in neural development, some of which we are functionally assessing in ongoing work.
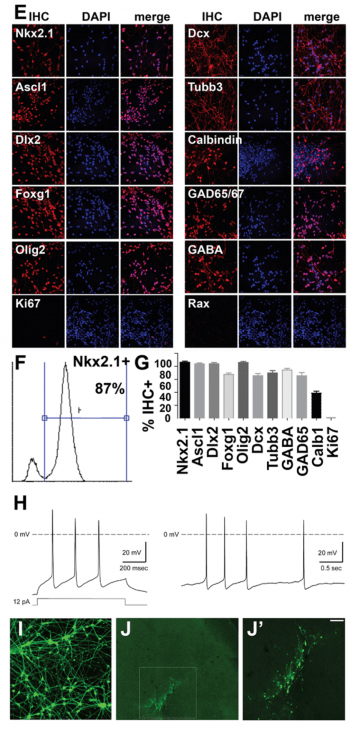
Cortical interneurons from human embryonic stem cells. (Figure E-J). Another major focus of our work is to define transcriptional networks and epigenetic regulatory controls underlying development of human cortical interneurons. The mammalian cerebral cortex consists of two major types of neuron, excitatory glutamatergic projection neurons that convey information to different regions of the brain and GABAergic interneurons that provide local inhibitory inputs to modulate responses of projection neurons and prevent over-excitation. Imbalances between excitatory and inhibitory neuronal activities can emerge during cortical development. These imbalances often reduce inhibitory signaling through dysfunctional specification, migration, differentiation, or survival of interneurons. Interneuron hypoplasia or dysfunction contributes to many neurological disorders, including epilepsy, schizophrenia, autism, and intellectual disability syndromes. Interestingly, after transplantation into the neonatal or adult nervous system, cortical interneurons have a striking ability to integrate into neural circuits and to provide inhibitory neuronal function in many locations, including their normal targets in the forebrain (hippocampus, cortex, striatum) as well as upon transplantation into heterologous locations such as the spinal cord. Because of this, transplanted interneurons acquired from the mouse medial ganglionic eminence have shown therapeutic utility in treating disease in mouse models of epilepsy, Parkinson’s disease, chronic pain, schizophrenia and anxiety.
It has recently become possible to generate mature cortical human interneurons from human embryonic stem cells (hESCs) and induced pluripotent stem cells (iPSCs). Genome engineering of hESC-derived interneurons enables us to study how genetic mutations lead to syndromes involving dysfunctional interneuron development. Using this approach, it may also be possible to develop human therapies akin to those used successfully to ameliorate several diseases in mouse models. Therefore, we are using these approaches: 1) to define how epigenetic regulation controls interneuron specification and differentiation, 2) to generate human mutations in epigenetic regulatory proteins that result in interneuron-based human disease, so that we can study how these mutations affect interneuron formation and function, and 3) to generate interneurons for cell transplantation-based approaches, which may have utility for treatment of disease states such as epilepsy and neuropathic pain.
Figure A-D. Gene regulatory network in embryonic neural progenitor cells (NPCs). Genomic binding profiles for seven transcription factors in embryonic NPCs were used (our Geminin ChIP-chip and ChIP-seq data and ref. 1-3, below). (A) Transcriptional and epigenetic regulatory activities that are direct targets of multiple nuclear regulatory transcription factors (Geminin direct targets highlighted) are shown. (B) Subset of the networked activities in A that cause neural tube defects (NTD) upon mutation in mouse models, (C-D) subset of the networked activities that show decreased expression either in embryonic (E10.5) neural tube tissue from Geminin conditional knockout mice (Mox2-Cre;CKO NTD model, ref. 4) or in E10.5 embryos from a diabetic pregnancy model of NTD susceptibility. E-J. Interneuron derivation from hESCs. Day 35 hESC-derived interneurons: (E) Immunohistochemistry (IHC) for ventral telencephalic/MGE, neuron, and interneuron markers. (F) FACS for Nkx2.1 and (G) quantitation of % IHC+ cells shown. (H) Representative action potential firing pattern. Left- evoked by injection of a square pulse, right- spontaeneous (data generated in collaboration with Jim Huettner). (I) Synapsin-GFP expressing day 35 interneurons (after PSA-NCAM+ selection) are (J-J’) engrafted into the cortex of P2 NOD/SCID mice (20 days post-engraftment shown; J’ boxed region in J, scale bar= 100µM) This can assess interneuron capacity for engraftment, migration, dispersion, and persistence in cortex and ability to ameliorate epilepsy phenotypes. References: 1. Bergsland, M., Ramsköld, D., Zaouter, C., Klum, S., Sandberg, R., & Muhr, J. (2011). Sequentially acting Sox transcription factors in neural lineage development. Genes & Development, 25(23), 2453–2464. doi:10.1101/gad.176008.111 2. Castro, D. S., Martynoga, B., Parras, C., Ramesh, V., Pacary, E., Johnston, C., et al. (2011). A novel function of the proneural factor Ascl1 in progenitor proliferation identified by genome-wide characterization of its targets. Genes & Development, 25(9), 930–945. doi:10.1101/gad.627811 3. Sansom, S. N., Griffiths, D. S., Faedo, A., Kleinjan, D.-J., Ruan, Y., Smith, J., et al. (2009). The level of the transcription factor Pax6 is essential for controlling the balance between neural stem cell self-renewal and neurogenesis. PLoS Genetics, 5(6), e1000511. doi:10.1371/journal.pgen.1000511 4. Patterson, E. S., Waller, L. E., & Kroll, K. L. (2014). Geminin loss causes neural tube defects through disrupted progenitor specification and neuronal differentiation. Developmental Biology. doi:10.1016/j.ydbio.2014.06.021 5. Pavlinkova, G., Salbaum, J. M., & Kappen, C. (2009). Maternal diabetes alters transcriptional programs in the developing embryo. BMC Genomics, 10, 274. doi:10.1186/1471-2164-10-274